The goal of our research is to combine insights into the biology, genetics, and molecular mechanisms of GIST and other sarcomas to develop novel therapeutics.
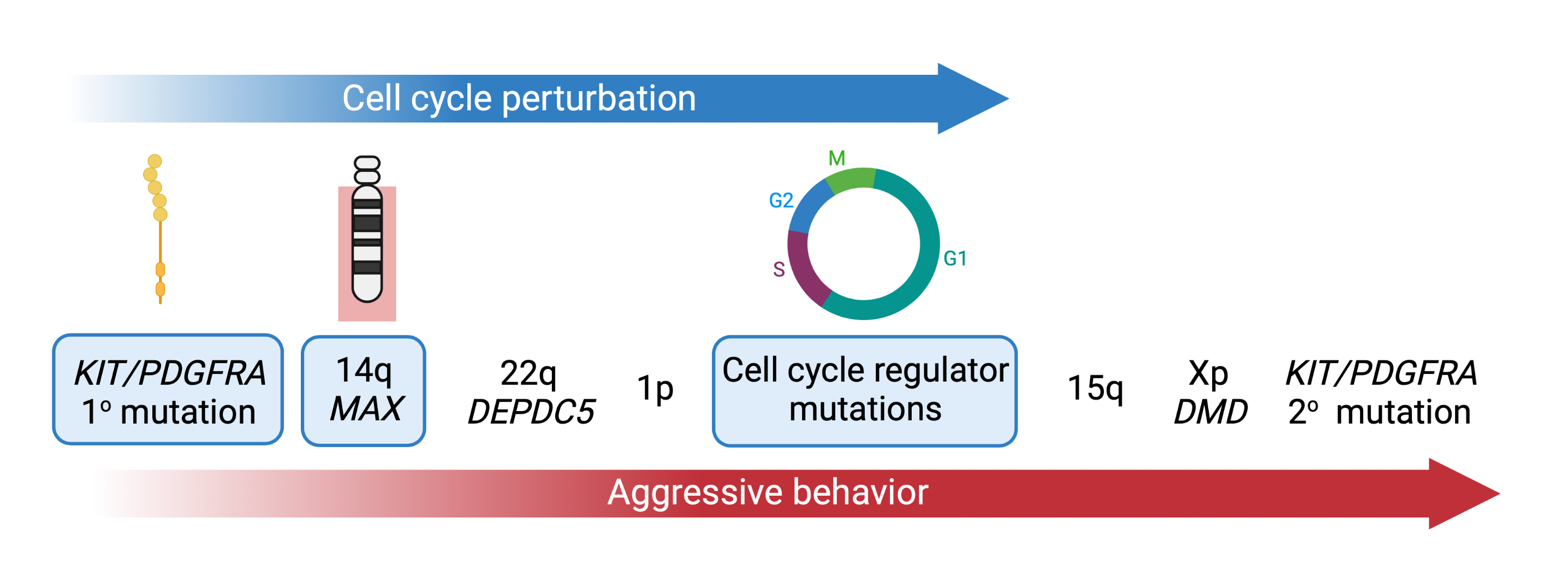
During malignant progression, GISTs acquire additional genomic hits such as deletions of 14q, 22q, 1p, or 15q. We showed that 14q deletions target the MAX tumor suppressor and lead to p16 silencing and early cell cycle dysregulation in GIST (Schaefer et al., Nature Communications. 2017). One focus of our group is to characterize the remaining steps in GIST progression.
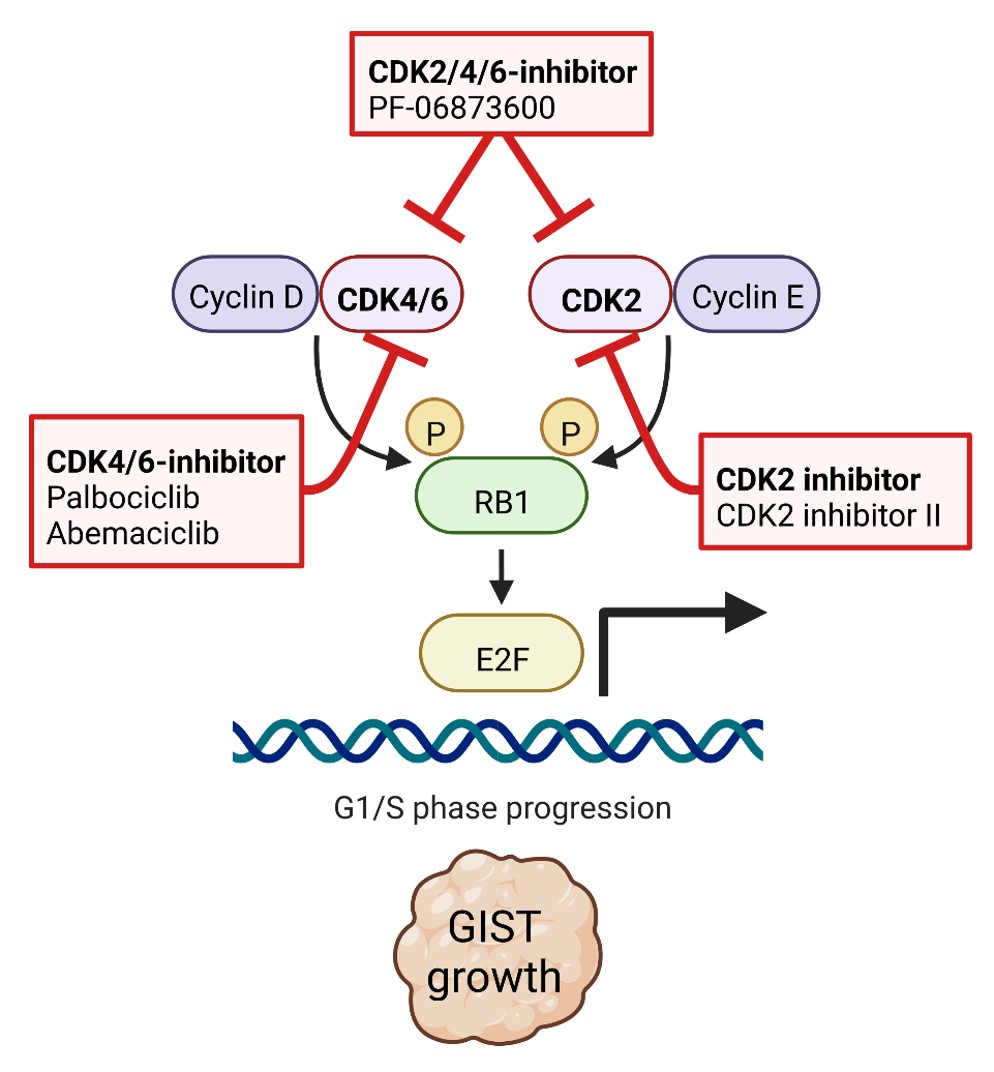
Our recent studies demonstrate that virtually all advanced GISTs are characterized by perturbations of the cell cycle machinery through genomic mutations that accumulate as GISTs progress. One focus of our group is to define the molecular mechanisms of cell cycle dysregulation in GIST, paving the way for the discovery of new diagnostic and therapeutic strategies using new inhibitor drugs (Schaefer et al., British Journal of Cancer. 2022).
The goal of our research is to translate these insights into other sarcoma types to define opportunities for therapeutic targeting. Our group demonstrated through protein expression screens that nearly all leiomyosarcomas (LMS) have aberrations of the key cell cycle regulators p53 or RB1, highlighting key roles of cell cycle perturbations in LMS development and progression (Schaefer et al., Cancer. 2021). This work provides a compelling rationale for therapies targeting these pathways. Our lab is interested in defining the role cell cycle aberrations in additional sarcoma types.
- PCR & Sanger sequencing
- RT-PCR
- Whole exome sequencing
- RNA sequencing
- CRISPR/Cas9 genome wide screens
- CRISPR/Cas9 gene knockout
- Cell-based assays
- Molecular methods
- Cell culture
- Xenograft studies
- Immunoblotting/Immunoprecipitation
- Immunohistochemistry
- Multiplexed IF
- Mass spectrometry